Abstract
Chimeric antigen receptor (CAR) T-cell therapies have revolutionized therapeutics for a variety of conditions. However, developing CAR T-cell therapies comes with many challenges, including costly cell engineering and culturing that require intensive and time-consuming manual processes. To address these challenges, Monomer Bio and Indee Labs teamed up to build an end-to-end automation system for producing CAR T-cell therapies. In this study, we evaluated the ability of this system, the Monomer Automation Platform, to increase culture throughput while maintaining T-cell health during culturing. We found that this system maintained T-cell proliferation, viability, total yield, and passage yield while increasing sample throughput by 8-fold. These findings support that the Monomer Automation Platform can effectively and efficiently culture and passage T cells—a critical step toward creating a powerful, automated system for developing CAR T-cell therapies for patients.
Introduction
Over the past decade, chimeric antigen receptor (CAR) T-cell therapies have revolutionized therapeutics for a variety of conditions, including cancer, heart disease, rare autoimmune diseases, and even aging.1 These remarkable therapies have shown long-term success, with some patients still in remission a decade after receiving treatment.2 Due to the success of CAR T-cell therapies, the Federal Drug Administration has approved 6 CAR T-cell therapies since 2017, and hundreds more are currently being tested in the clinic.3,4 Although this rapid growth is promising, the process of designing, producing, and testing CAR T-cell therapies comes with many challenges.
One challenge of CAR T-cell therapies involves difficulties in the rigorous development process. CAR T-cell therapies are made by collecting blood samples from patients, extracting peripheral blood mononuclear cells (PBMCs) from these samples, and then isolating the T cells from the population of PBMCs. These isolated T cells are then engineered to express a CAR protein complex that will target specific types of cells, such as cancer cells. After the engineered CAR T-cells are tested, they are infused into patients as a therapeutic agent.5
In this development process, 2 notable difficulties arise.5,6 First, the engineering step has historically relied on lentiviral and retroviral delivery mechanisms to efficiently modify host-cell DNA to introduce the CAR gene. However, this approach is costly and requires important biosafety considerations for large-scale production and testing.7 Second, the T cells must be cultured and expanded before and after the engineering step. This culturing process requires daily attention over many weeks, and any errors can lead to expensive delays that last weeks or even months.
To address these difficulties, we teamed up with Indee Labs to build an end-to-end automation system, or workcell, for producing CAR T-cell therapies. The workcell will eventually include an instrument called the Hydropore, developed by Indee Labs, that efficiently delivers DNA, RNA, CRISPR (clustered regularly interspaced short palindromic repeats) Cas9 ribonucleoproteins, and more to cells via transfection.8,9 The workcell is controlled by our Monomer Automation Platform, which includes software that enables fully automated T-cell culturing and engineering.
In this study, we evaluated the ability of our Monomer Automation Platform to increase culture throughput while maintaining T-cell health before the T-cell engineering step.
Materials & Methods
T-cell Population
Primary T cells were enriched from human PBMCs (STEMCELL Technologies, CAT# 72005) using the EasySep Human T Cell Isolation Kit (STEMCELL Technologies, CAT# 17591).
Samples from the same donor were cultured with the manual and automated methods (described below) at the same time.
Manual Cell Culture
Cell-culture timeline. On day 0, fresh T cells were seeded in a 12-well plate with culture media. On day 1, a sample was collected from each well and run on a flow cytometer to determine the cell count. Fresh media was then added to each well as described below. This process was repeated on day 4. On day 7, a sample was collected from each well and run on a flow cytometer to determine the cell count. Then the cells were passaged to a new 12-well plate as described below. On day 8, a sample was collected from each well of the new plate and run on a flow cytometer to determine the cell count.
Media additions. A 12-well plate that contained T cells growing in culture media (X-VIVO-20 + 1% human serum) was removed from the Inheco SCILA incubator (95% air, 5% CO2, 37℃, 85%-95% relative humidity). A cell count was determined with a flow cytometer (see the Flow Cytometry section). Media addition volumes were manually calculated for each well of the 12-well culture plate as follows:

The culture media was warmed in a 37ºC water bath for 30 minutes or until the culture media reached 37ºC (measured using a JVTIA JT-320 temperature gun). The warmed media and cell-culture plate were transferred to a sterile hood. Media was added to each well of the plate as previously calculated. Each well was mixed by aspirating and dispensing the culture slurry 2 times with a P300 pipette. The cell-culture plate was then placed in the Inheco SCILA incubator.
Cell passaging. A 12-well plate that contained T cells growing in culture media (X-VIVO-20 + 1% human serum) was removed from the Inheco SCILA incubator (95% air, 5% CO2, 37℃, 85%-95% relative humidity). A cell count was determined with a flow cytometer (see the Flow Cytometry section). A cell-aliquot volume was manually calculated for each well as follows:
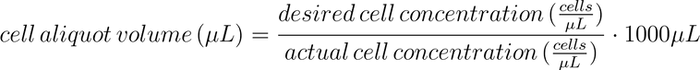
Culture media was warmed in a 37ºC water bath for 30 minutes (or until the culture media reached 37ºC measured using a JVTIA JT-320 temperature gun). The warmed media, cell-culture plate, and an aliquot of interleukin-2 (IL-2) were transferred to a sterile hood. Each well of the cell-culture plate was gently mixed by twice aspirating and dispensing the cell slurry with a P300 pipette head. The calculated cell-aliquot volume of cell slurry was extracted from each well and dispensed 1:1 into a new well of a new 12-well plate. Fresh media was added to each well in the new plate to bring the total volume up to 1000 µL. IL-2 (10 µL/1 mL media) was added to each well in the new plate, and each well was mixed by gently aspirating and dispensing the cell slurry 3 times with a P300 pipette. The cell-culture plate was then placed in the Inheco SCILA incubator.
Automated Cell Culture
Cell-culture timeline. On day 0, fresh T cells were seeded in a 24-well plate with culture media. On day 1, a sample was collected from each well and run on a flow cytometer to determine the cell count. Fresh media was then added to each well as described below. This process was repeated on day 4. On day 7, a sample was collected from each well and run on a flow cytometer to determine the cell count. Then the cells were passaged to a new 24-well plate as described below. On day 8, a sample was collected from each well of the new plate and run on a flow cytometer to determine the cell count.
Workcell setup. Automation experiments were performed on the Indee Labs T-cell culture workcell built by Monomer Bio (Figure 1). Media additions and passages were performed on the Opentrons OT-2 liquid handler. Media was prewarmed on the Opentrons Heater/Shaker module. Culture plates were incubated in the Inheco SCILA incubator, and reagent plates were stored in a mini-fridge outfitted with a custom sliding door. A plate hotel was used to store plates at room temperature. Plates were moved between instruments and locations by the KX-2 robotic arm. In a 2-mL deep 96-well plate, 1.2 mL of culture media was added to each well using an 8-channel manual pipette (“Media Plate”). In a second 2-mL deep 96-well plate, 1 mL of an IL-2:media solution (55 µL IL-2/1 mL media) was added to each well (“Cytokine Plate”). Both plates were stored at approximately 4℃.

Indee Labs’ researchers used the Monomer Automation Platform software to carry out the automated experiments.
Media additions. A 24-well plate containing T cells growing in culture media (X-VIVO-20 + 1% human serum) was removed from the incubator (95% air, 5% CO2, 37℃, 85%-95% relative humidity). A cell count was determined with a flow cytometer (see Flow Cytometry section). Then the culture plate was returned to the incubator. Media addition volumes were manually calculated as follows:

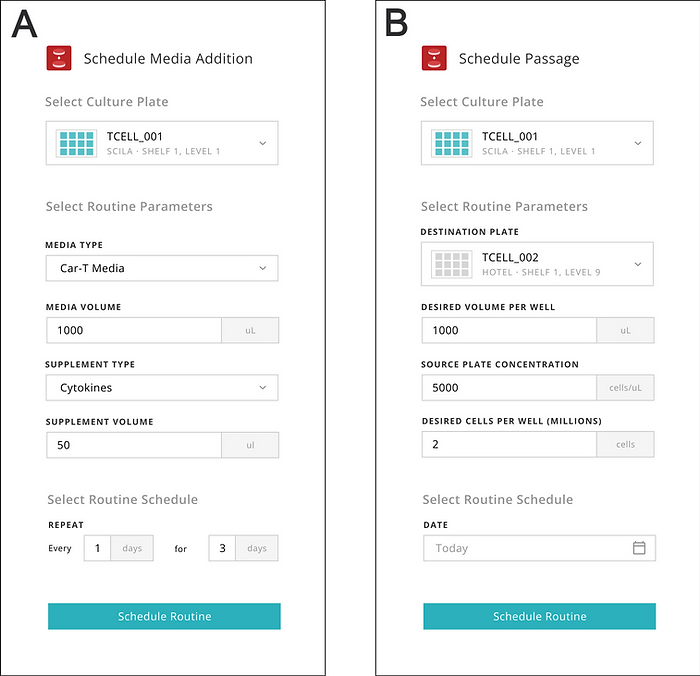
The Indee Labs operator used the Monomer software interface to enter the calculated volumes and schedule the media additions (Figure 2). From this point, the remaining work was fully performed by the workcell controlled by the Monomer Automation Platform software.
The robotic arm opened the refrigerator door, removed the Media Plate, and moved the Media Plate to the Heater/Shaker module on the OT-2 deck. The Media Plate was warmed on the module for 30 minutes at 37℃. The tray holding the lidded cell-culture plate in the incubator was automatically opened, and the culture plate was moved from the incubator to the liquid-handling deck by the robotic arm. The lids of the culture and media plates were removed by the robotic arm. Then the OT-2 dispensed the appropriate volume of media, sourced from the Media Plate, into each well of the cell-culture plate one well at a time using a P300 tip. The Monomer software tracked consumption of the Media Plate and ensured each addition was performed successfully (ie, the OT-2 did not try to aspirate media from an empty well of the Media Plate). Once the OT-2 finished adding the media to each well, the robotic arm replaced the lid on the plate and moved it back to the incubator.
Cell passaging. A 24-well plate that contained T cells growing in culture media (X-VIVO-20 + 1% human serum) was removed from the Inheco SCILA incubator (95% air, 5% CO2, 37℃, 85%-95% relative humidity). A cell count was determined with a flow cytometer (see Flow Cytometry section). Then the culture plate was returned to the incubator. A cell-aliquot volume was manually calculated for each well as follows:
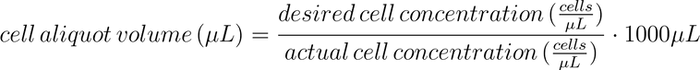
A new 24-well plate was placed onto the plate hotel. The Indee Labs operator used the Monomer software interface to enter the cell-aliquot volumes and the new plate’s location, and to schedule the passages (Figure 2). From this point, the remaining work was fully performed by the workcell controlled with the Monomer Automation Platform software.
The robotic arm opened the refrigerator door, removed the Media Plate, and moved the Media Plate to the Heater/Shaker module on the OT-2 deck. The Media Plate was warmed on the module for 30 minutes at 37℃. The new 24-well plate was then moved from the plate hotel to the OT-2 deck. The robotic arm then opened the refrigerator door, removed the Cytokine Plate, and moved the Cytokine Plate to the OT-2 deck. The robotic arm removed the lids from the Media and Cytokine Plates. Then the OT-2 dispensed media from the Media Plate into each well of the new 24-well plate using a P1000 tip. The amount of media dispensed was enough to bring the total volume up to 1000 µL. Then the OT-2 dispensed 50 µL Cytokine solution, sourced from the Cytokine Plate, into each well of the new 24-well plate. The robotic arm then replaced the lid on the Media and Cytokine Plates and placed them back at approximately 4℃.
Next, the tray holding the lidded cell-culture plate in the incubator was automatically opened, and the culture plate was moved from the incubator to the liquid-handling deck by the robotic arm. The lid was removed from the culture plate by the robotic arm. Each well of the culture plate was gently mixed using a compass pattern method that Monomer Bio developed on the OT-2. After mixing, the OT-2 aspirated the appropriate cell aliquot volume for the given well from the source culture plate and dispensed that volume into a new 24-well plate in the matching well location where the cells were resuspended in the previously dispensed media. This approach ensured that all wells were re-seeded at the same density. The robotic arm then replaced the lid on the new 24-well plate and moved the plate to the incubator. Then the source culture plate was discarded.
Flow Cytometry
Cell count. Cell counts were determined using AutoCount, a method Monomer Bio and Indee Labs developed on the Thermo Fisher Attune Flow Cytometer. Each well of the culture plate to be sampled was removed from the incubator and mixed by aspirating and dispensing the slurry 2 times with a pipette (manual experiments: 12-well plate; automated experiments: 24-well plate). Then 50 µL of cell slurry was removed from each well of the culture plate and dispensed into a 96-well plate row by row starting at position A1. The sample plate was inserted into the Attune, and the AutoCount method was run. Raw results were processed in the Attune software to produce the count. The concentration of the sample was calculated (cell count/sample volume). The cell count for each well of the original culture plate was then estimated (concentration total well volume).
Viability. Cell viability was measured using the Thermo Fisher Attune Flow Cytometer. Each well of the culture plate to be sampled was from the incubator and mixed by aspirating and dispensing the slurry 2 times with a pipette (manual experiments: 12-well plate; automated experiments: 24-well plate). Then 50 µL of cell slurry was removed from each well of the culture plate and dispensed into a new 96-well plate row by row starting at position A1. A propidium iodide stain was loaded in the plate, and the plate was inserted into the Attune, and the viability assay was run. Raw results were processed in the Attune software to produce a viability percentage for the sample.
Calculations. The cell expansion rate was calculated by converting the cell concentration recorded from the cell-count assay to an estimated count for the source well (concentration × volume). The fold change in cell count for each well was plotted over time. The cell yield was calculated as the number of viable cells on day 7 of expansion (total cell count % viability). The passaging yield was calculated as the number of viable cells 1 day after passaging (total cell count % viability).
Results
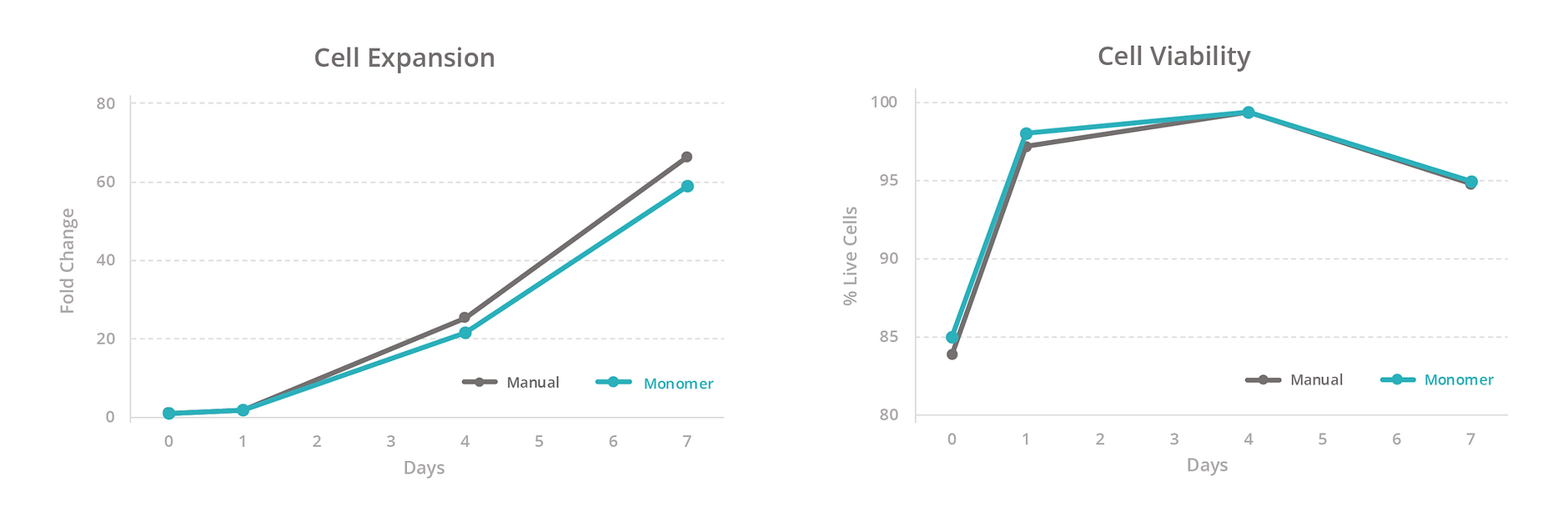
To evaluate our platform’s ability to maintain T-cell health, we compared the growth rate, viability, and total yield of primary human T cells expanded manually versus with the Monomer Automation Platform. We found that T cells cultured manually or with the automated platform had similar growth rates and viability at all measured timepoints (Figure 3), as well as similar total yield after 7 days of culturing (Figure 4). Also, T cells passaged manually or with the automated platform produced similar passaging yields and viability 1 day after passaging (Figure 5).
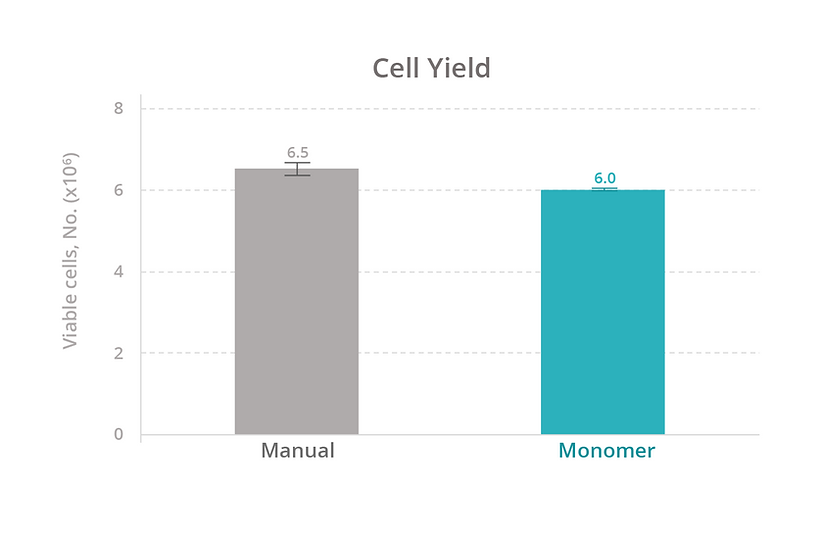
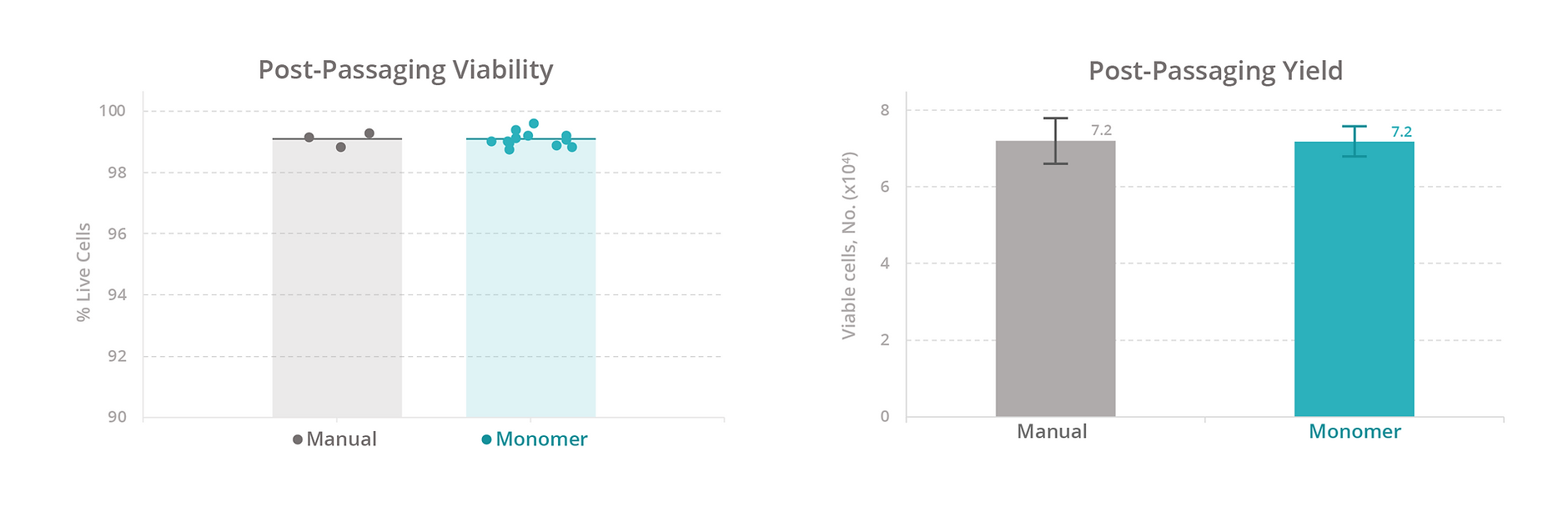
We also compared the efficiency of maintaining cells manually versus with the automated platform. With manual culturing, a simple media exchange takes an average of 15 minutes of active time to process 12 samples. With our automated platform, a media exchange takes approximately 15 minutes to set up a media exchange on 96 samples (four 24-well plates at the same time). Based on these estimates, the automated platform increases sample throughput by 8-fold (data not shown).
Discussion
In this study, we evaluated the ability of the Monomer Automation Platform to maintain T-cell health while increasing sample throughput. We found that the automated platform maintained T-cell proliferation, viability, total yield, and passage yield similar to the manual method. The automated platform also increased sample throughput by 8-fold compared to the manual method. These findings support the value of using the Monomer Automation Platform to effectively and efficiently culture and passage T cells.
To further streamline the automated process, we designed the software for the Monomer Automation Platform to include additional time-saving benefits. For example, the software supports advanced scheduling and remote monitoring of media changes and cell passages (Figure 2). These features reduce the time needed in the laboratory and eliminate the need for managing cell cultures in person during weekends.
We are also taking steps to further refine the platform. Our next step is to fully automate the process for preparing for and executing the cell-count assay. With this automation, we will be able to automate the media addition and passaging calculations so that the system can dynamically count, feed, and passage cells according to the real-time state of the cells. By working with the real-time state of the cells, we can process cells when they are ready to be acted on with the automated platform rather than process them when the scientist is ready to act on the cells. When culturing by hand, scientists often passage a plate all at once, even if some wells are still at relatively low concentration (ie, not ready to be acted on). The Monomer Automation Platform allows us to passage each well when it reaches a specific concentration, which reduces well-to-well variability and minimizes the total number of passage events while maximizing cell yield. Also, when completed manually, the count and passage steps take 3 hours to complete. Our goal is to use the automated platform to reduce that time to only 15 active minutes. We also plan to integrate a larger, automated cell-culture incubator to further increase sample capacity and throughput.
Another important feature of an end-to-end automation system for engineering T cells is the Hydropore. The Hydropore has already been used to successfully permeabilize cell membranes to incorporate foreign material and enable gene editing. This platform is also compatible with high-throughput screening workflows.9,10 Eventually, by combining the Hydropore with the Monomer Automation Platform, we will create a powerful, automated system for culturing and engineering T cells.
In conclusion, we showed that the Monomer Automation Platform can efficiently and effectively culture and passage T cells. These preliminary findings bring us one step closer to fully automating the T-cell engineering process. Although we will continue to refine the Monomer Automation Platform, our successful collaboration with Indee Labs will create a powerful, automated system for designing and producing CAR T-cell therapies for patients.
References
- Zmievskaya E, Valiullina A, Ganeeva I, Petukhov A, Rizvanov A, Bulatov. Application of CAR-T cell therapy beyond oncology: autoimmune diseases and viral infections. Biomedicines. 2021;9(1):59. doi:10.3390/biomedicines9010059
- Melenhorst JJ, Chen GM, Wang M, et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature. 2022:602(7897):503-509. doi:10.1038/s41586-021-04390-6
- Moreno-Cortes E, Forero-Forero JV, Lengerke-Diaz PA, Castro JE. Chimeric antigen receptor T cell therapy in oncology – pipeline at a glance: analysis of the ClinicalTrials.gov database. Crit Rev Oncol Hematol. 2021;159:103239. doi:10.1016/j.critrevonc.2021.103239
- Maalej KM, Merhi M, Inchakalody VP. CAR-cell therapy in the era of solid tumor treatment: current challenges and emerging therapeutic advances. Mol Cancer. 2023;22(1):20. doi:10.1186/s12943-023-01723-z
- Levine BL. Performance-enhancing drugs: design and production of redirected chimeric antigen receptor (CAR) T cells. Cancer Gene Ther. 2015;22(2):79-84. doi:10.1038/cgt.2015.5
- CAR T cells: engineering immune cells to treat cancer. National Cancer Institute. Updated March 10, 2022. Accessed Month DD, YYYY. https://www.cancer.gov/about-cancer/treatment/research/car-t-cells
- Harris E, Elmer JJ. Optimization of electroporation and other non‐viral gene delivery strategies for T cells. Biotechnol Prog. 2021;37(1):e3066. doi:10.1002/btpr.3066
- Jarrell JA, Twite AA, Lau KHWJ, et al. Intracellular delivery of mRNA to human primary T cells with microfluidic vortex shedding. Sci RepI. 2019;9(1):1-11. doi:10.1038/s41598-019-40147-y
- Jarrell JA, Sytsma BJ, Wilson LH, et al. Numerical optimization of microfluidic vortex shedding for genome editing T cells with Cas9. Sci Rep. 2021;11(1):1-13. doi:10.1038/s41598-021-91307-y
- Kasper SH, Otten S, Squadroni B, et al. A high-throughput microfluidic mechanoporation platform to enable intracellular delivery of cyclic peptides in cell-based assays. Bioeng Transl Med. 2023;8(15):e10542. doi:10.1002/btm2.10542




